Allogeneic cell therapies are attractive to cell therapy developers over autologous cell therapies for many reasons. One blood cell donation or cord blood unit can be manufactured into many doses. The therapy is available off the shelf when a patient needs it. And allogeneic cell therapies could potentially be made at a lower cost per patient than an autologous therapy. Not to mention the huge potential for clinical research provided by an available off-the-shelf product.
Many cell therapy developers also believe they can overcome the human leukocyte antigen (HLA) barrier. While cell therapy developers may be able to edit their way around some HLA barriers, the issue of histocompatibility should not be overlooked.
HLA isn’t the only histocompatibility factor to consider
HLA matching is the aspect of histocompatibility that people are usually the most familiar with.
Allogeneic hematopoietic stem cell transplantation (HSCT) requires HLA matching to reduce the likelihood of graft-versus-host disease, in which the donor cells recognize the patient’s cells as foreign and create an immune attack. In HSCT, we match on four or more clinically relevant HLA genes. Those that impact clinical outcomes are HLA-A, B, C, DRB1 and DQB1.
However, there is a histocompatibility aspect that is extremely important, but sometimes overlooked by cell therapy developers. That is the concept of antibody histocompatibility.
Patients can be synthesized against donor-specific antibodies (DSAs). These pre-formed antibodies in the patient are directed against the donor’s class I (HLA-A, B, C) or class II (DRB1, DQB1, DRB3/4/5, DPB1) HLA antigens and beyond. If a patient has DSAs, they could reject the donor cells.
We use a process called cross-match to test for compatibility. Cross-match can be completed in a lab, however, virtual cross-match is becoming a more common way to screen for DSAs. Virtual cross-match is the process of assessing the results of solid phase and cell-based HLA antibody identification assays to predict, or correlate to, the results of a physical cross-match.
Screening for DSAs is extremely important, particularly in the context of HLA-mismatched HSCTs or HLA-dependent allogeneic cellular therapies to avoid early graft rejection. When the donor and patient are HLA-matched, particularly for the clinically relevant HLA loci, it may be beneficial to investigate DSAs targeting HLA loci that were not accounted for in the match process (e.g., DPA1, DQA1 or DRB3/4/5).
With more than 500 allogeneic cell therapies in the pipeline, cell therapy developers must consider the impact histocompatibility could have on patient outcomes.
Other considerations for allogeneic cell therapy donor selection
Histocompatibility is an important consideration for allogeneic cellular therapy development. However, other concepts are also critical in donor selection and the design of an off-the-shelf cell bank. One of those is the concept of HLA geographical ancestry and ancestral haplotypes.
A haplotype is a group of alleles that fall on the same chromosome. HLA genes are very tightly linked on the short arm of chromosome 6 and are therefore mostly inherited in blocks. HLA genes encode the human immune system that is shaped over generations by numerous selective pressures exerted on humans depending on which area of the world they live in.
This leads to the concept of ancestral haplotypes that are specific to certain areas of the world. A haplotype that is very common in Southern Asia may not be common in Latin America, for example. These ancestral haplotypes travel through generations.
Using HSCT as an example, a patient with a certain HLA type is more likely to be a full match with a donor whose ancestors came from the same area of the world because they are more likely to have similar ancestral haplotypes. The concept of ancestral haplotypes is represented in our systems by estimating HLA haplotype frequencies from the HLA of registry members belonging to different populations. Each population has a different set of haplotype frequencies.
At NMDP℠, haplotype frequency information allows our matching algorithm, HapLogic℠, to apply imputation for missing and ambiguous data. The more than 41 million potential donors on worldwide registries don’t all have the same level of HLA typing information available. Those who have been on a registry longer will have less information available because typing technology has advanced over time.
In this simplified example, HapLogic identified three potential matches for a patient. There are some gaps in the data for the three donors. The patient is Asian. The three potential donors are Asian, African American and European American. HapLogic used the haplotype frequency information to calculate the match likelihood. In this scenario, the donor who is Asian is more likely to be a full match for the patient.
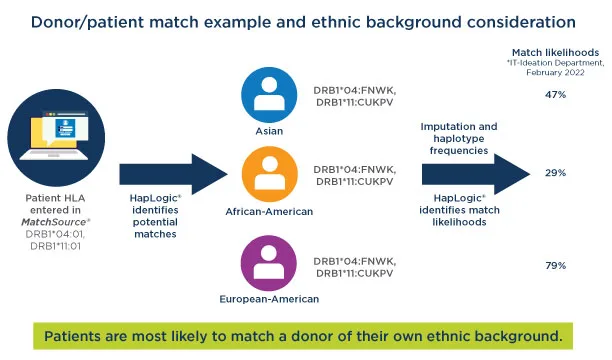
Haplotype frequency impact on off-the-shelf cell bank design
This ancestral haplotype information is extremely useful when modeling an allogeneic cell bank, too.
Cell therapy developers often ask our NMDP BioTherapies℠ Bioinformatics Consulting team, “What is the optimal off-the-shelf cell bank design?” We can use the information we have on donors and cord blood units—including haplotype, KIR, CMV status, gender, etc.—and our unmatched expertise to create the model.
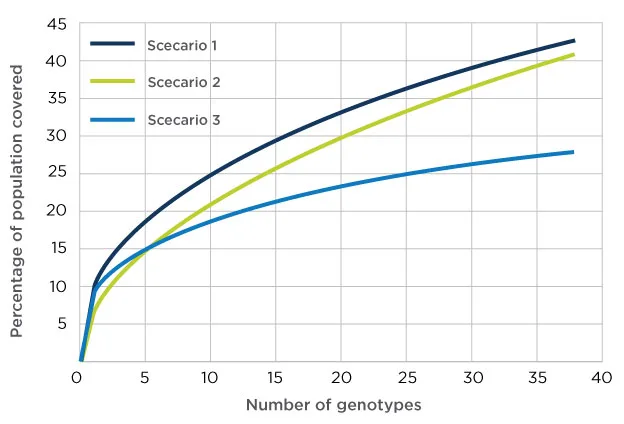
Data-driven insights for your allogeneic cell therapy
Our NMDP BioTherapies Bioinformatics Consulting team has unique experience that allows us to develop methods that support proper donor/patient histocompatibility for allogeneic cell therapies.
Explore how our data-driven insights can help you accelerate your allogeneic cell therapy development and manage risk.





